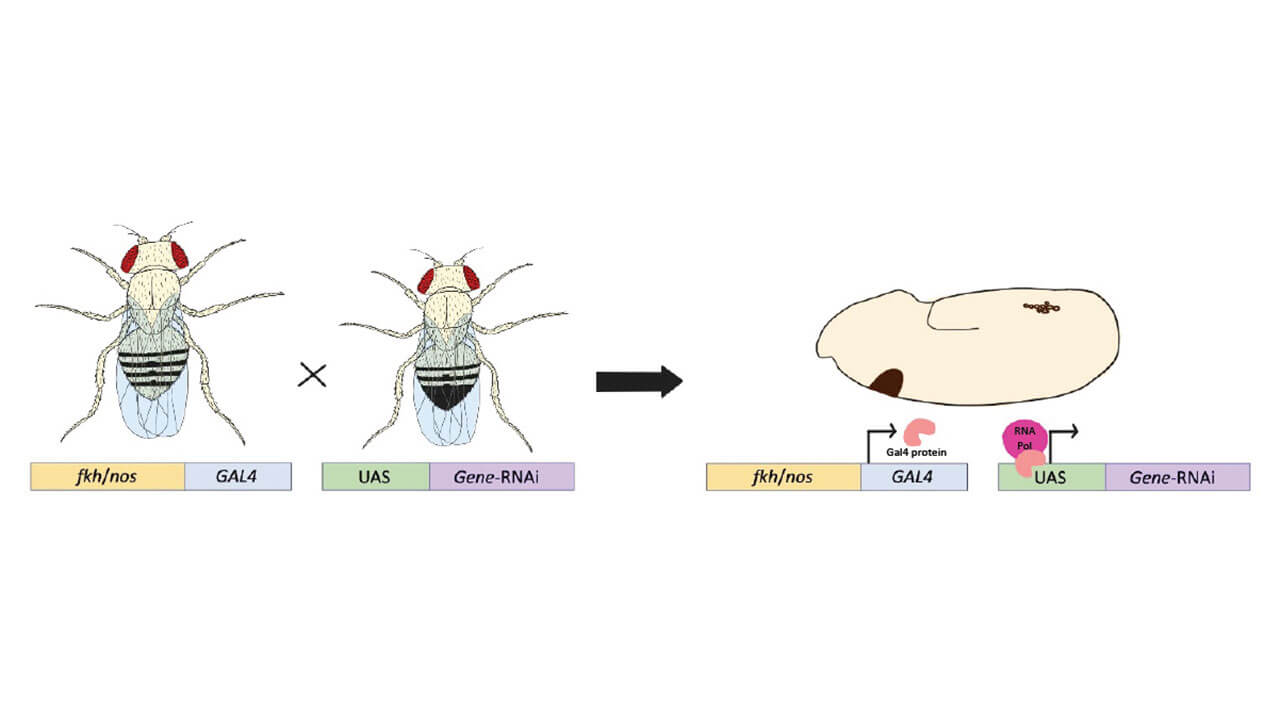
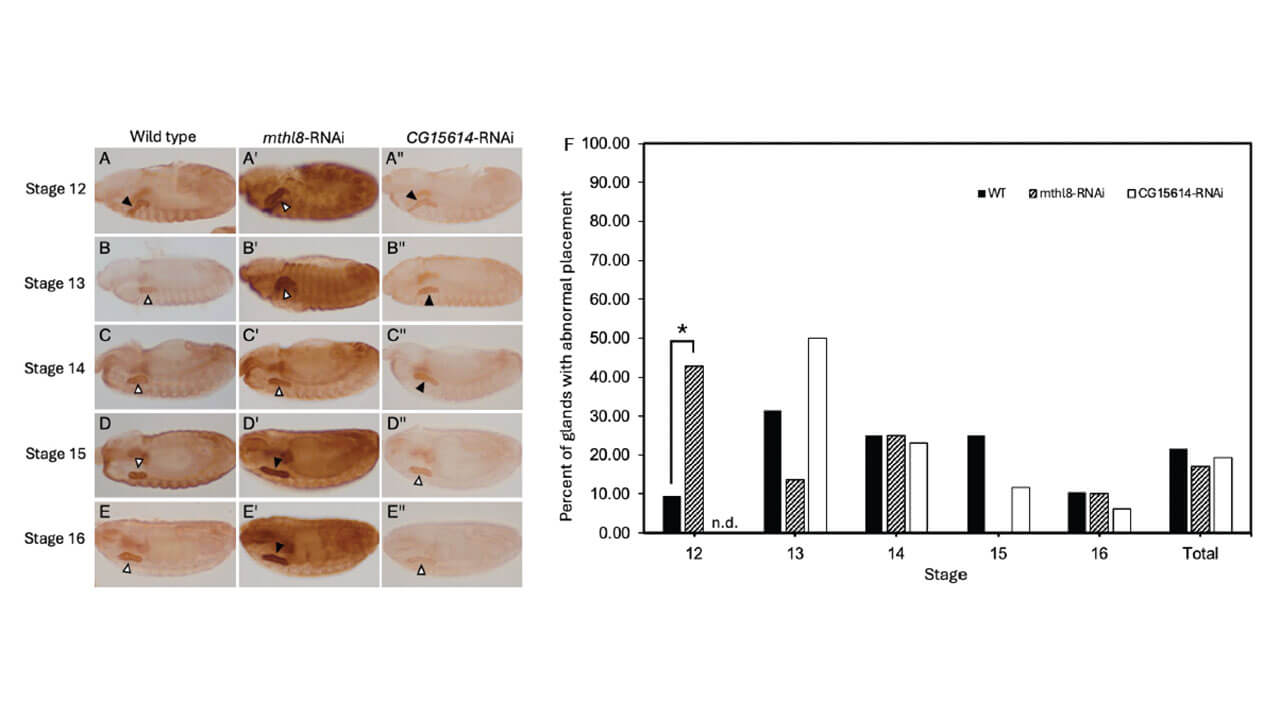
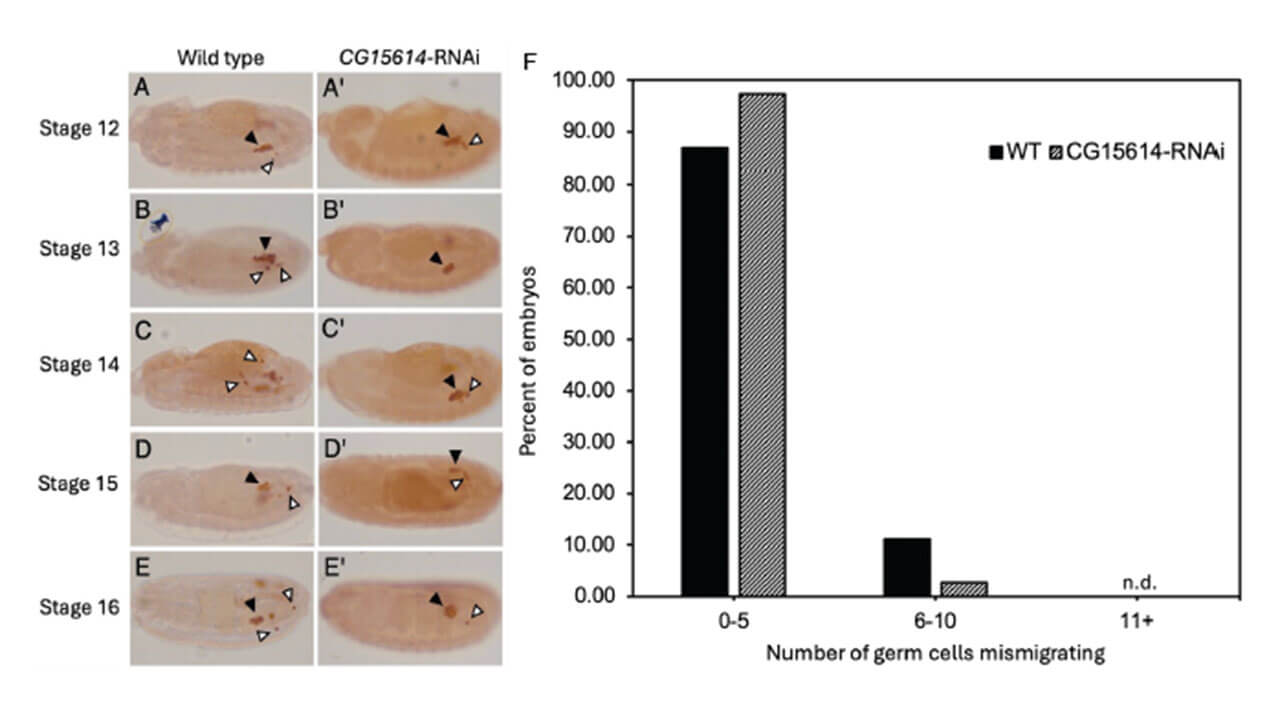
Professional Application
”I was looking for research experiences to better prepare me for both medical school and a job in the research industry. When I heard about QUIP-RS, I knew it was a perfect opportunity to expand on this interest, and even better with someone I’m close to as my mentor. I gained a lot more insight into the field of scientific research than I was anticipating and fell in love with the process more than I thought; I’ve gone from an aspiring MD to an aspiring MD/PhD after this program! There is still much to learn, but I’m so grateful this was my first real taste of research.“ - Hailey Tolson ’25
Associate Professor of Biology
Biological Sciences
For Further Discussion
This serves as an overview of the project and does not include the complete work. To further discuss this project, please email Hailey Tolson.
Quinnipiac University Interdisciplinary Program for Research and Scholarship
Open to students of all majors, QUIP-RS provides up to $5,000 in funding for undergraduate students to conduct research or complete creative projects alongside faculty mentors. This intensive 8-week program enables students to develop scholarly skills while encouraging discussion about successes and shortcomings with fellows and mentors.
Learn more about QUIP-RS
Explore Our Areas of Interest
We've sorted each of our undergraduate, graduate and doctoral programs into unique Areas of Interest. Explore these categories to discover which programs and delivery methods best align with your educational and career goals.
Explore Physical and Life Sciences at Quinnipiac
Explore STEM Programs at Quinnipiac
References
Fredriksson, R. and Schiöth, H. B. 2005. The Repertoire of G-Protein-Coupled Receptors in Fully Sequenced Genomes. Mol. Pharm. 67(5): 1414-1425. 10.1124/mol.104.009001
Gramates, L. S., Agapite, J., Attrill, H., Calvi, B. R., Crosby, M. A., Dos Santos, G., Goodman, J. L., Goutte-Gattat, D., Jenkins, V. K., Kaufman, T., Larkin, A., Matthews, B. B., Millburn, G., Strelets, V. B. 2022. FlyBase: a guided tour of highlighted features. Genetics 220(4). iyac035.
Hanlon, C. D. and Andrew, D. J. 2015. Outside-in signaling – a brief review of GPCR signaling with a focus on the Drosophila GPCR family. J. Cell Sci. 128(19): 3533-3542. 10.1242/jcs.175158
Hu Y, Comjean A, Rodiger J, Liu Y, Gao Y, Chung V, Zirin J, Perrimon N, Mohr SE. FlyRNAi.org-the database of the Drosophila RNAi screening center and transgenic RNAi project: 2021 update. Nucleic Acids Res. 2021 Jan 8;49(D1):D908-D915. doi: 10.1093/nar/gkaa936. PMID: 33104800; PMCID: PMC7778949.
Kolesnikov, T. and Beckendorf, S. K. 2005. NETRIN and SLIT guide salivary gland migration. Dev. Bio. 284(1): 102-111. 10.1016/j.ydbio.2005.04.037
Ribeiro, C., Petit, V., Affolter, M. 2003.
Signaling systems, guided cell migration, and organogenesis: insights from genetic studies in Drosophila. Dev. Bio. 260(1): 1-8. doi.org/10.1016/S0012-1606(03)00211-2
Warrington, S. J., Strutt, H., Strutt, D. 2013. The Frizzled-dependent planar polarity pathway locally promotes E-cadherin turnover via recruitment of RhoGEF2. Development. 140(5): 1045-1054. http://doi.org/10.1242/dev.088724
Williamson, A. and Lehmann, R. 1996. Germ Cell Development in Drosophila. Annu. Rev. Cell Dev. Biol. 12: 365-391. 10.1146/annurev.cellbio.12.1.365