QUIP-RS
Structural Variation in Spina Bifida Patient Genomes

QUIP-RS
Structural Variation in Spina Bifida Patient Genomes
The goal of this study conducted by Kaylee Pettengill ’27 was to uncover CNVs using whole-genome sequencing data to investigate the affected biological processes relating to the altered neural tube fusion.
Overview
This research employed the use of bioinformatics with whole-genome sequencing data from proband-parent trios to uncover unbalanced structural variants that potentially implicate the development of spina bifida in the fetus through altered neural tube fusion and related biological processes.
Researcher

Kaylee Pettengill '27
Biomedical Sciences
School of Health Sciences
Identifying Structural Variation in Spina Bifida Genomes
Abstract
Spina bifida arises from incomplete closure of the neural tube during early gestation of the embryo and is a complex condition leading to neurological and functional impairments (Iskandar & Finnell, 2022). Structural variants (SVs) of the genome, including copy number variants (CNVs), may contribute to the developmental mechanisms that result in this condition. Our goal was to uncover CNVs using whole-genome sequencing (WGS) data to investigate the affected biological processes relating to the altered neural tube fusion. Two computational scripts were developed and deployed for this process: the first to call CNVs, and the second to filter the identified variants and to identify variants conferring risk. Visual validation was conducted to ensure high-confidence findings. Our analyses identified 14 high-impact CNVs that may be associated with spina bifida. These variants were further examined to understand their inheritance patterns relating to the proband’s parents and to investigate expression pattens. Our identified CNVs suggest potential developmental and functional implications for neural tube fusion during gestation.
Introduction
Spina bifida is a congenital condition caused by incomplete fusion of the neural tube, which forms the central nervous system during the third week of gestation. This defect leaves the spinal cord exposed without protective coverings like dura, bone, muscle, or skin, making the fetus vulnerable to infection and cerebrospinal fluid. Spina bifida often leads to significant complications such as leg, bladder, and bowel dysfunction, as well as nerve damage, resulting in the need for specialized, long-term care in adulthood1.
Structural variation, which includes quantitative and structural rearrangements of chromosomes, plays a major role in genetic diversity and is especially relevant in the study of rare diseases, cancer genetics, and evolutionary genetics2. Structural variants (SVs), such as copy number variants (CNVs), can range from 50 base pairs to entire chromosomes and may impact gene expression and dosage in ways that contribute to developmental disorders3. This project aims to screen for structural variation using whole-genome sequencing data from proband-parent trios to investigate their role and contribution to spina bifida.
Materials and Methods
To investigate copy number variations (CNVs) potentially implicated in the development of spina bifida, we analyzed data from 89 case-parent trios in our cohort, comprising over 5.8 TB of genomic data. Structural variants were identified using CNVnator and ERDS, tools that leverage read-depth analysis (CNVnator for variable call sizes and ERDS for incorporating SNP data). The analyses yielded approximately 12,267 CNV calls. We filtered these calls based on multiple criteria, including predicted loss-of-function intolerance (pLI) scores and allele frequency, to determine the variants with the highest potential impact. Selected CNVs were visually validated using Integrated Genomics Viewer (IGV) and Samplot, which resulted in a final call set of 14 rare, high-impact, genic CNVs.
Results
The 14 identified CNVs impacted genes with diverse functions, loci, and sizes. Notably, all the affected genes were expressed in the central nervous system, with several also expressed in the fetal brain. This expression pattern suggests a potential role for these CNVs in disrupting biological pathways critical to neural tube development, thereby contributing to the formation of spina bifida during the early stages of embryonic growth. Several of the identified CNVs were novel and not previously reported in genomic databases.
This circos plot depicts a genome-wide representation of CNVs detected in the study.
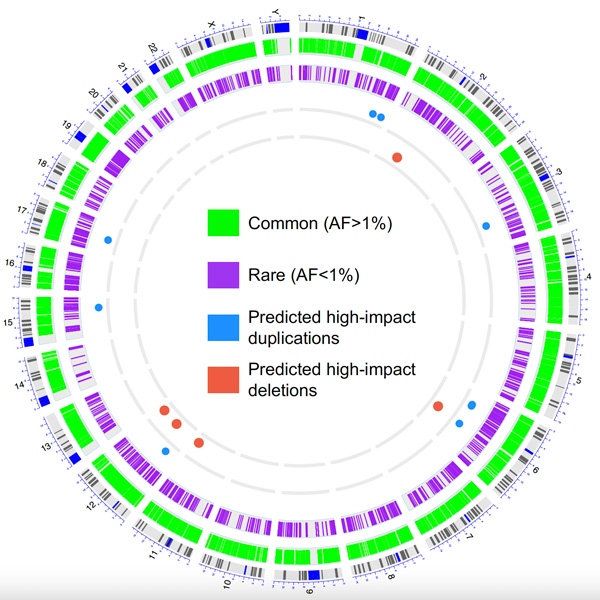
Conclusions
Through the analysis of whole-genome sequencing data from spina bifida probands and their unaffected parents, we identified a variety of structural variants, including 14 copy number variants (CNVs) most likely implicated in the development of spina bifida that were inherited from the non-affected parents. Notably, some of these CNVs have never been previously recorded in genomic databases, suggesting novel contributions to the condition. All 14 CNVs are expressed in tissues critical for neural development, including the brain and spinal cord, with many specifically expressed in embryonic and fetal development. These findings underscore the potential functional roles that rare CNVs may play in neural tube fusion.
Future Directions
Future directions include:
- Functional investigations of these implicated CNVs and genes using CRISPR genome editing in model organisms to directly observe developmental impacts
- Conducting statistical transmission disequilibrium tests for both SNVs and SVs in our cohorts
- Characterization of balanced structural variants and other cohorts of various ethnicities and population admixtures to refine our findings
Professional Application
"This project has prepared me with valuable skills for a career in healthcare by challenging me to learn, adjust and build confidence in coding and script development — skills I previously had no experience with. The ability to adapt is a necessary skill for the fast-paced environment and complex tasks physicians are faced with in every setting. I thoroughly enjoyed the intellectually stimulating and logic-based thinking required to devise solutions for the complex problems I faced. This project has not only contributed to genetic and spina bifida research, but also to my growth and development as a student and future healthcare professional." – Kaylee Pettengill ’27
Faculty Mentor
For Further Discussion
This serves as an overview of the project and does not include the complete work. To further discuss this project, please email Kaylee Pettengill.
Quinnipiac University Interdisciplinary Program for Research and Scholarship
Open to students of all majors, QUIP-RS provides up to $5,000 in funding for undergraduate students to conduct research or complete creative projects alongside faculty mentors. This intensive 8-week program enables students to develop scholarly skills while encouraging discussion about successes and shortcomings with fellows and mentors.
Explore Our Areas of Interest
We've sorted each of our undergraduate, graduate and doctoral programs into unique Areas of Interest. Explore these categories to discover which programs and delivery methods best align with your educational and career goals.
Explore Health and Medicine Programs at Quinnipiac
Explore Physical and Life Sciences at Quinnipiac
Explore STEM Programs at Quinnipiac
References
[1] Iskandar, B. J., & Finnell, R. H. (2022). Spina bifida. N Engl J Med, 387(5), 444-450. doi:10.1056/NEJMra2116032
[2] Spielmann, M., Lupiáñez, D. G., & Mundlos, S. (2018). Structural variation in the 3D genome. Nature Reviews Genetics, 19(7), 453-467
[3] NewYork-Presbyterian. (n.d.). Image of Pediatric Orthopedics Services [Image]. NewYork-Presbyterian. https://www.nyp.org/pediatrics/orthopedics
[4] Overview of Structural Variation [Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; 2004 – [cited 2024 March 8]. Available from: https://www.ncbi.nlm.nih.gov/gene/
